Polymerase Chain Reaction (PCR) is a highly sensitive and specific technique used in clinical laboratories to amplify small segments of DNA. It is widely used for detecting genetic mutations, diagnosing infectious diseases, and performing genetic research.
- Principle: PCR relies on temperature-dependent enzymatic cycles to amplify DNA or RNA.
- Non-Culture-Based: It doesn’t require culturing cells; instead, it directly amplifies nucleic acids.
- Cyclic Process: Through multiple cycles of denaturation, annealing, and extension, PCR quickly generates billions of copies of the target DNA or RNA segment.
- Hybridization and Replication: PCR combines nucleic acid hybridization and replication principles. It starts by denaturing double-stranded DNA, then selects a segment for amplification using specific primers, followed by replication with DNA polymerase.
- Origin: PCR was developed in the mid-1980s by Kary Mullis and his team. Mullis and Michael Smith were awarded the Nobel Prize in Chemistry in 1993 for this groundbreaking innovation.
- Revolutionary Impact: PCR has become a cornerstone in molecular biology and genetics, enabling DNA and RNA analysis with profound implications for research, diagnostics, and various fields.

Key Points:
Polymerase Chain Reaction (PCR) is a revolutionary technique in molecular biology that allows for the amplification of a specific DNA sequence. Here are some key points about PCR:
- Amplification: PCR rapidly multiplies a specific DNA sequence.
- Heating and Cooling: It involves cycles of heating to separate DNA strands and cooling to allow primers to bind.
- Targeted: Amplifies a specific region of DNA.
- Enzyme: DNA polymerase enzyme synthesizes new DNA strands.
- Applications: Used in research, diagnostics, forensics, and more.
- Versatility: Can detect pathogens, identify genetic disorders, and analyze DNA samples.
- Speed: Provides results in a matter of hours.
- Sensitivity: Detects even small amounts of DNA.
- Variants: Includes qPCR, RT-PCR, nested PCR, multiplex PCR, and dPCR.
- Revolutionary: Revolutionized molecular biology and genetics.
Introduction
Polymerase Chain Reaction (PCR) is a highly sensitive and specific technique used in clinical laboratories to amplify small segments of DNA. It is widely used for detecting genetic mutations, diagnosing infectious diseases, and performing genetic research.
Types of PCR
Several modification of PCR methods have been developed to enhance the utility of this method in diagnostic settings based on their applications. Some of the common types of PCR are;
- Real-Time PCR
- Nested PCR
- Multiplex PCR
- Quantitative PCR
- Arbitrary Primed PCR
Principle
PCR is based on the enzymatic replication of DNA. It involves repeated cycles of denaturation, annealing, and extension to amplify a specific DNA sequence exponentially. Taq polymerase, a heat-stable DNA polymerase, is used to synthesize new DNA strands.
Specimen Requirements
- Type of specimen: Blood, serum, plasma, tissue, swabs, or other body fluids
- Volume of specimen required: Typically 50-200 µL for blood, less for other samples
- Collection method: Standard venipuncture for blood, swabbing for mucosal samples
- Handling and storage requirements: Store samples at 2-8°C for short-term or -20°C or lower for long-term storage
- Stability of the specimen: DNA is stable for several days at 2-8°C and for months to years at -20°C or lower
Reagents and Materials
- DNA extraction kit or reagents
- PCR master mix (containing Taq polymerase, dNTPs, MgCl2, and buffer)
- Forward and reverse primers specific to the target DNA sequence
- Nuclease-free water
- Positive control DNA template
- Negative control (no DNA template)
- PCR tubes or plates
- Mineral oil (if using a thermal cycler without a heated lid)
Equipment
- Thermal cycler (PCR machine)
- Micropipettes and tips
- Vortex mixer
- Centrifuge
- Agarose gel electrophoresis apparatus (for post-PCR analysis)
- UV transilluminator or gel documentation system (for visualizing PCR products)
Procedure
DNA Extraction:
- Extract DNA from the specimen using a commercial DNA extraction kit or standard extraction protocols.
- Quantify and assess the purity of the extracted DNA using a spectrophotometer or fluorometer. PCR Setup:
- Prepare the PCR master mix according to the manufacturer’s instructions. The typical components per reaction include:
- 10-50 ng of template DNA
- 0.2-0.5 µM of each primer
- 1X PCR buffer
- 1.5-2.5 mM MgCl2
- 200 µM dNTPs
- 0.5-1.0 U of Taq polymerase
- Nuclease-free water to the final reaction volume (usually 20-50 µL)
- Aliquot the master mix into PCR tubes or a 96-well plate.
- Add the template DNA to the tubes or wells containing the master mix.
- Include a positive control with a known DNA template and a negative control without DNA. Thermal Cycling:
- Place the tubes or plate in the thermal cycler.
- Program the thermal cycler with the following cycling conditions (typical example):
- Initial denaturation: 94°C for 2-5 minutes
- 30-40 cycles of:
- Denaturation: 94°C for 30 seconds
- Annealing: 50-65°C for 30 seconds (depending on primer Tm)
- Extension: 72°C for 30-60 seconds (depending on amplicon length)
- Final extension: 72°C for 5-10 minutes
- Start the PCR run. Post-PCR Analysis:
- Prepare an agarose gel (e.g., 1-2% agarose in TAE or TBE buffer).
- Load the PCR products and a DNA ladder into the gel wells.
- Run the gel at an appropriate voltage until the dye front has migrated sufficiently.
- Stain the gel with ethidium bromide or SYBR Safe.
- Visualize the PCR products under a UV transilluminator or gel documentation system.
Quality Control
- Use positive and negative controls to validate each PCR run.
- Ensure all reagents and consumables are free from contamination.
- Verify the specificity and efficiency of primers through proper validation steps.
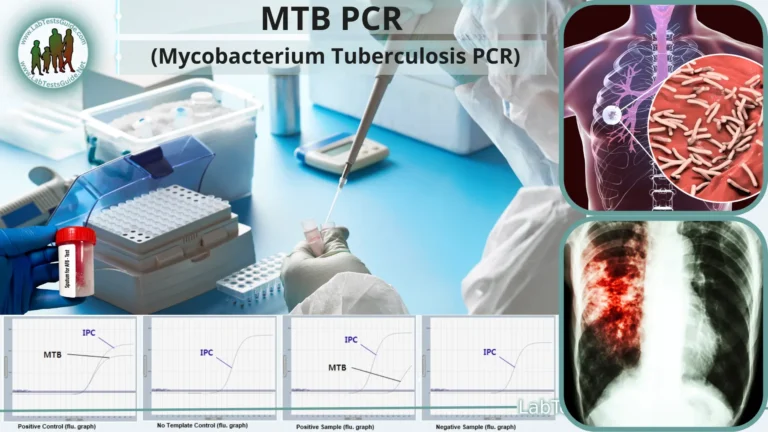
Calculation and Interpretation
- Compare the size of the PCR products with the DNA ladder to confirm the expected product size.
- Analyze the presence or absence of bands to determine the presence of the target DNA sequence in the samples.
- Consider potential sources of error, such as contamination or non-specific amplification.
Limitations
- PCR is susceptible to contamination, leading to false positives.
- Inhibitors in clinical samples can affect the efficiency of the PCR.
- High GC content or secondary structures in the template can impede amplification.
Safety Precautions
- Wear appropriate PPE (gloves, lab coat, safety goggles).
- Handle all reagents and samples according to standard biohazard protocols.
- Use separate areas for pre-PCR and post-PCR procedures to prevent contamination.
- Dispose of waste according to local regulations.
References
- Mullis, K., & Faloona, F. (1987). Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol, 155, 335-350.
- Clinical Chemistry: Principles, Techniques, and Correlations by Michael L. Bishop, Edward P. Fody, and Larry E. Schoeff.
- Tietz Textbook of Clinical Chemistry and Molecular Diagnostics.
- Manufacturer’s instructions for PCR reagents and thermal cyclers.
- Clinical Laboratory Standards Institute (CLSI) guidelines.
This format can be adapted for various types of PCR, such as real-time PCR (qPCR) or reverse transcription PCR (RT-PCR), by adjusting the specific details to match the methodology used for each type of assay.