Dot blotting is a molecular biology technique used to detect and analyze specific proteins or nucleic acids in a sample. It is a simplified variation of the more widely known technique called Western blotting (immunoblotting) used for protein detection. Dot blotting is particularly useful when you have limited quantities of a sample or when you want a quicker, less labor-intensive method compared to Western blotting.
Key Points of Dot Blotting:
- Purpose: Dot blotting is a molecular biology technique used for the qualitative detection of specific proteins, DNA, RNA, or other molecules in a sample.
- Sample Types: Dot blotting can be applied to a wide range of sample types, including proteins, nucleic acids, and other biomolecules.
- Simplified Technique: It is a simplified alternative to Western blotting and is often used when you have limited sample material.
- Sample Application: Small volumes of the sample are directly applied to a solid support membrane, typically made of nitrocellulose or nylon.
- Immobilization: The samples are allowed to air-dry or can be dried using a vacuum, immobilizing the target molecules on the membrane.
- Blocking: After drying, the membrane is blocked to prevent non-specific binding of detection reagents.
- Blocking Buffer: Blocking is typically done using a blocking buffer containing proteins like BSA or milk.
- Probe Binding: A specific probe, such as an antibody or complementary nucleic acid probe, is applied to the membrane. This probe will bind to the target molecules if they are present in the sample.
- Washing: Unbound probe and blocking solution are removed by washing the membrane.
- Detection Methods: Dot blotting can employ various detection methods, including colorimetric, chemiluminescent, or fluorescent techniques, depending on the probe used.
- Visualization: The presence of the target molecule is indicated by the appearance of a signal (e.g., color change or light emission) at the location of the dot.
- Qualitative Analysis: Dot blotting provides qualitative rather than quantitative data about the presence or absence of a target molecule.
- Reduced Sensitivity: It may have reduced sensitivity compared to other techniques like ELISA or quantitative PCR (qPCR).
- Cross-Reactivity: Non-specific binding or cross-reactivity with other molecules can lead to false-positive results.
- Rapid Screening: Dot blotting is suitable for rapid screening of a large number of samples simultaneously.
- Limited Quantification: It is less quantitative compared to Western blotting, as the signal intensity does not directly correlate with the amount of the target molecule.
- Versatility: Despite its limitations, dot blotting remains a valuable tool for initial screenings and qualitative assessments of target molecules in samples.
Defination of Dot Blotting:
Dot blotting is a molecular biology technique used for the qualitative detection of specific proteins, DNA, RNA, or other molecules in a sample by applying small drops or “dots” of the sample onto a solid support membrane and probing it with specific molecules (e.g., antibodies or nucleic acid probes) to identify the presence or absence of the target molecule.
Background and Significance:
Background History:
- Development in the 1970s: Dot blotting emerged as a simplified alternative to more complex techniques like Western blotting and Southern blotting in the 1970s.
- Nucleic Acid Origins: Initially, dot blotting was primarily used for the detection of nucleic acids (DNA and RNA).
- Protein Adaptation: Over time, it was adapted for protein detection, becoming a versatile tool for molecular biology.
Significance:
- Sample Conservation: Dot blotting is significant for its ability to detect target molecules in samples with limited material, preserving precious samples.
- Simplicity and Speed: It offers a rapid and straightforward method for detecting specific molecules compared to more intricate techniques.
- Screening Tool: Dot blotting is valuable for screening large numbers of samples quickly, making it an essential tool in research and diagnostics.
- Qualitative Assessment: It provides qualitative information about the presence or absence of target molecules in a sample, aiding in initial assessments.
- Cost-Effective: Dot blotting is often more cost-effective than other quantitative assays, making it accessible to a wide range of laboratories.
- Versatility: It can be adapted for various biomolecules, including proteins, DNA, RNA, and more, making it versatile in different research contexts.
- Limited Resources: Especially important in resource-limited settings, where complex equipment and reagents may not be readily available.
- Historical Role: Dot blotting played a historical role in the development of molecular biology techniques, paving the way for modern molecular diagnostics.
Purpose of Dot Blotting:
- Qualitative Detection: Dot blotting is primarily used for the qualitative detection of specific molecules (proteins, DNA, RNA) in a sample.
- Limited Sample Material: It is valuable when you have limited sample material and cannot afford to use it for more resource-intensive techniques.
- Initial Screening: Dot blotting serves as an initial screening tool to determine the presence or absence of target molecules in a sample.
- Rapid Analysis: It offers a quicker and simpler alternative to techniques like Western blotting, Southern blotting, or Northern blotting.
- Preservation: Dot blotting conserves samples by requiring only a small amount for analysis.
- Versatility: It can be adapted for various types of biomolecules, making it versatile in different research areas.
- Cost-Effective: Dot blotting is often more cost-effective than quantitative assays, making it accessible to a wide range of laboratories.
- Resource-Limited Settings: It is useful in settings where access to complex equipment and reagents is limited.
- Confirmation of Results: Dot blotting can be used to confirm the presence of specific molecules identified through other assays.
- Molecular Diagnostics: It has applications in diagnostic tests for diseases and genetic disorders.
- Protein-Protein Interactions: Dot blotting can be used to study protein-protein interactions by immobilizing one protein and probing for interactions with another.
- Environmental Analysis: In environmental science, dot blotting can be used to detect specific contaminants or microorganisms in environmental samples.
- Pharmacological Studies: It is employed in drug discovery and pharmacological studies to assess the presence of specific drug targets.
- Comparative Analysis: Researchers use dot blotting to compare levels of specific molecules between different samples or conditions.
- Quality Control: Dot blotting can be used in quality control processes, such as assessing the purity of recombinant proteins.
- Antibody Screening: It is useful in antibody development and validation by assessing the specificity of antibodies.
- Viral Detection: Dot blotting can be applied in viral detection and the study of viral proteins.
- Genetic Research: In genetics, dot blotting is used to detect and study DNA methylation patterns and genetic mutations.
- Protein Expression: Researchers employ dot blotting to evaluate protein expression levels in cells or tissues.
- Biomarker Discovery: It aids in the identification of potential biomarkers associated with diseases or physiological processes.
- Gene Expression Profiling: Dot blotting can be adapted for gene expression studies, providing insights into RNA levels.
- Food Safety: In food science, dot blotting can be used for detecting foodborne pathogens or allergenic proteins in food samples.
Applications of Dot Blotting:
- Protein Detection: Dot blotting is used to detect specific proteins in a sample, aiding in protein identification and characterization.
- Nucleic Acid Detection: It can detect DNA or RNA molecules, making it useful in genetic research and diagnostic testing.
- Antibody Screening: Dot blotting is employed to screen and validate antibodies for their specificity in recognizing target antigens.
- Viral Detection: It can be used to detect viral proteins or nucleic acids in clinical samples, contributing to viral diagnostics.
- Biomarker Discovery: Dot blotting helps identify potential biomarkers associated with diseases or conditions for diagnostic and research purposes.
- Gene Expression Profiling: It aids in studying gene expression by detecting specific RNA molecules in samples.
- Environmental Monitoring: Dot blotting can be applied in environmental science to detect specific contaminants or microorganisms in environmental samples.
- Drug Discovery: Researchers use dot blotting to assess the presence of specific drug targets in biological samples.
- Quality Control: It is used in quality control processes, such as assessing the purity and identity of recombinant proteins.
- Comparative Analysis: Dot blotting is valuable for comparing levels of specific molecules between different samples or experimental conditions.
- Pharmacological Studies: It plays a role in pharmacological studies to evaluate the presence of specific molecular targets in tissues or cells.
- Disease Diagnostics: Dot blotting is employed in clinical diagnostics to detect disease-related biomolecules, aiding in disease diagnosis and prognosis.
- Immunological Research: It is used to study immune responses, antigen-antibody interactions, and the presence of specific antigens in biological samples.
- Food Safety: In food science, dot blotting is used for detecting allergenic proteins or foodborne pathogens in food samples, ensuring food safety.
- Genetic Mutations: Researchers apply dot blotting to detect specific genetic mutations or variations in DNA sequences.
- DNA Methylation Analysis: It aids in the study of DNA methylation patterns, which can be relevant in epigenetics and cancer research.
- Cell Signaling Studies: Dot blotting can be used to detect specific signaling molecules in cell lysates or tissue extracts.
- Microbiological Research: In microbiology, dot blotting can be used to identify and study specific microorganisms or microbial components.
- Tumor Marker Detection: It helps in detecting tumor-specific markers in cancer research and clinical diagnostics.
- Protein-Protein Interaction Studies: Researchers immobilize one protein and probe it for interactions with other proteins, facilitating the study of protein-protein interactions.
- Drug Resistance Testing: It can be employed to detect specific mutations associated with drug resistance in pathogens.
- Autoimmune Disease Research: Dot blotting can be used to identify autoantibodies associated with autoimmune diseases.
Principles of Dot Blotting:
- Sample Application: Dot blotting begins with the application of a small volume (a “dot”) of the sample directly onto a solid support membrane. Common membrane materials include nitrocellulose, nylon, or PVDF (polyvinylidene fluoride).
- Immobilization: The applied sample is allowed to dry on the membrane or is dried using a vacuum, resulting in the immobilization of the target molecules from the sample onto the membrane surface.
- Blocking: To prevent non-specific binding of detection reagents (e.g., antibodies or nucleic acid probes), the membrane is blocked with a blocking buffer. Common blocking agents include bovine serum albumin (BSA) or non-fat milk.
- Probe Binding: A specific probe, such as an antibody for protein detection or a complementary nucleic acid probe for DNA/RNA detection, is applied to the membrane. The probe is designed to bind specifically to the target molecule if it is present on the membrane.
- Incubation: The membrane is incubated with the probe, allowing for specific binding between the probe and the target molecule.
- Washing: Unbound probe and excess blocking solution are removed by washing the membrane with a buffer, leaving only the bound probe behind.
- Detection: The presence of the target molecule is detected through various methods, depending on the type of probe used. Common detection methods include colorimetric, chemiluminescent, or fluorescent techniques.
- Visualization: The appearance of a signal (e.g., color change, luminescence, or fluorescence) at the location of the dot on the membrane indicates the presence of the target molecule.
- Qualitative Analysis: Dot blotting provides qualitative data, indicating whether the target molecule is present or absent in the sample. It does not provide quantitative information about the concentration of the molecule.
- Comparison: The results can be compared between different samples or conditions, helping researchers assess the relative presence or absence of the target molecule.
- Limitations: Dot blotting has limitations, including reduced sensitivity compared to quantitative techniques, the potential for cross-reactivity, and limited quantification capabilities.
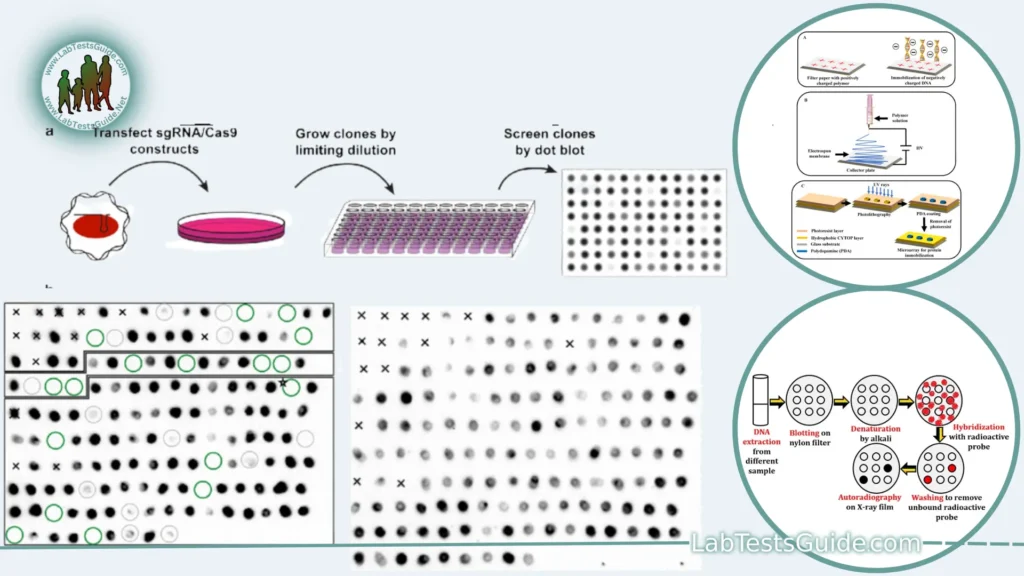
Procedure for Dot Blotting:
- Sample Preparation: Prepare your sample containing the target molecules (e.g., proteins, DNA, RNA).
- Membrane Placement: Place a solid support membrane (e.g., nitrocellulose or nylon) on a flat surface.
- Sample Application: Using a micropipette, apply small drops or “dots” of your sample directly onto the membrane.
- Drying: Allow the dots to air-dry or use a vacuum-assisted method to dry the sample drops, immobilizing the target molecules on the membrane.
- Blocking: Incubate the membrane in a blocking buffer (e.g., BSA or milk) to prevent non-specific binding.
- Probe Application: Apply a specific probe (e.g., antibody or nucleic acid probe) that binds to the target molecule you want to detect.
- Incubation: Incubate the membrane with the probe, allowing for specific binding.
- Washing: Wash the membrane to remove unbound probe and excess blocking solution.
- Detection: Use an appropriate detection method (e.g., colorimetric, chemiluminescent, or fluorescent) to visualize the presence of the target molecule.
- Signal Visualization: The appearance of a signal (e.g., color change, luminescence, or fluorescence) at the location of the dot indicates the presence of the target molecule.
- Qualitative Assessment: Interpret the results qualitatively, determining whether the target molecule is present or absent.
- Comparison: Compare the results between different samples or conditions, if applicable.
Materials and Reagents:
Materials:
- Solid Support Membrane: (e.g., nitrocellulose or nylon membrane) for immobilizing the sample and probe.
- Micropipette: For precise application of sample and reagents.
- Vacuum Apparatus: Optional but useful for speeding up drying of sample dots on the membrane.
- Flat Surface: To support the membrane during the procedure.
- Blocking Buffer: (e.g., BSA or non-fat milk) to prevent non-specific binding of detection reagents.
- Specific Probe: (e.g., antibody or nucleic acid probe) designed to bind to the target molecule.
- Washing Buffer: For removing unbound probe and excess blocking solution.
- Detection System: (e.g., substrate for colorimetric detection, chemiluminescent reagents, or fluorescent detection system) to visualize the presence of the target molecule.
Reagents:
- Sample: Contains the target molecules you want to detect (e.g., protein extract, DNA, RNA).
- Primary Antibody: If detecting proteins, a specific primary antibody is needed for binding to the target protein.
- Secondary Antibody: If using a primary antibody, a secondary antibody labeled with a detection molecule (e.g., enzyme, fluorophore) may be required for signal amplification.
- Lysis Buffer: If detecting intracellular proteins, a lysis buffer is used to extract proteins from cells or tissues.
- Tween-20: Often added to washing buffers to reduce non-specific binding.
- Salts and Buffers: Various salts and buffer solutions (e.g., Tris-buffered saline) for preparing buffers and maintaining appropriate pH levels.
- Chemicals for Blocking: If using a specific blocking agent, such as BSA or non-fat milk, these chemicals are required.
- Substrate: Specific substrates may be needed for colorimetric or chemiluminescent detection.
Step-by-Step Protocol:
- Prepare Membrane:
- Cut the solid support membrane to the desired size.
- Place the membrane on a flat surface (e.g., petri dish).
- Sample Application:
- Prepare your sample containing the target molecules. Ensure proper dilution if needed.
- Using a micropipette, apply small drops (dots) of your sample onto the membrane. Arrange dots in rows or any desired pattern.
- Drying: Allow the membrane to air-dry at room temperature. Alternatively, use a vacuum apparatus to speed up the drying process. Ensure that the dots are fully dry.
- Blocking: Incubate the membrane in blocking buffer for 1-2 hours at room temperature or overnight at 4°C. This step helps prevent non-specific binding.
- Primary Antibody (for Protein Detection):
- If detecting proteins, dilute the primary antibody in blocking buffer according to the manufacturer’s recommendations.
- Incubate the membrane with the primary antibody solution for 1-2 hours at room temperature or overnight at 4°C. This allows the primary antibody to bind to the target proteins.
- Secondary Antibody (if using primary antibody):
- If using a primary antibody, dilute the secondary antibody (conjugated with a detection molecule) in blocking buffer.
- Incubate the membrane with the secondary antibody solution for 1-2 hours at room temperature. This step amplifies the signal.
- Washing: Wash the membrane with washing buffer (TBST) 3-4 times for 5-10 minutes each to remove unbound antibodies.
- Detection:
- Prepare the detection reagents (e.g., chemiluminescent substrate) according to the manufacturer’s instructions.
- Apply the detection reagent onto the membrane for the appropriate duration, typically a few minutes.
- Signal Visualization: Visualize the presence of the target molecule by observing the appearance of a signal (e.g., luminescence or color change) at the location of the dots on the membrane.
- Qualitative Assessment: Interpret the results qualitatively to determine whether the target molecule is present or absent based on the appearance of the signal.
- Data Recording: Record and document the results, including images if necessary.
- Comparison (Optional): Compare the dot blot results between different samples or experimental conditions.
Result Interpretation:
Interpreting the results of a dot blot involves assessing the presence or absence of the target molecule based on the appearance of signals or dots on the membrane. Here’s a general guide for result interpretation in dot blotting:
- Positive Result:
- A positive result indicates the presence of the target molecule in the sample. This is typically indicated by the appearance of one or more distinct dots or signals on the membrane.
- The intensity of the signal may vary, with stronger signals indicating a higher concentration of the target molecule.
- Negative Result:
- A negative result suggests the absence of the target molecule in the sample. In this case, there are no visible dots or signals on the membrane.
- Controls:
- To validate your results, include positive and negative controls in your dot blot experiment.
- The positive control should be a sample known to contain the target molecule, and it should produce a positive signal.
- The negative control should be a sample known to lack the target molecule, and it should not produce any signal.
- Comparing your sample results to these controls helps confirm the accuracy of your dot blot.
- Qualitative Assessment:
- Dot blotting provides qualitative rather than quantitative data. You can assess the presence or absence of the target molecule but not its exact concentration.
- Results should be interpreted based on the visual appearance of the dots or signals and their comparison to controls.
- Image Analysis (Optional):
- If needed, you can use image analysis software to quantitatively assess the signal intensity. This can provide semi-quantitative data but is less accurate than other quantitative techniques.
- Confirmation:
- If the results are critical or if further quantification is required, consider confirming the findings using more quantitative techniques like Western blotting (for protein detection) or quantitative PCR (for nucleic acid detection).
- Replication:
- It’s advisable to replicate your dot blot experiments to ensure the reliability of your results.
- Documentation:
- Record and document the results, including images if captured, to maintain a clear record of your experiments.
- Troubleshooting:
- If the results are unexpected or unclear, consider troubleshooting steps such as optimizing the blocking conditions, changing antibody concentrations, or confirming the quality of your antibodies.
- Data Interpretation:
- Interpret the results in the context of your research question or hypothesis. The presence or absence of the target molecule may have implications for your study.
Remember that dot blotting is a qualitative technique primarily used for initial screenings and assessments. It provides a “yes” or “no” answer regarding the presence of the target molecule but does not quantify its exact concentration. If precise quantification is required, more quantitative methods should be employed.
Advantages and Disadvantages of Dot Blotting:
Advantages of Dot Blotting:
- Simplicity: Dot blotting is a straightforward and easy-to-understand technique, making it accessible to a wide range of researchers.
- Rapid Results: It provides relatively quick results compared to more time-consuming methods like Western blotting or PCR.
- Limited Sample Quantity: Dot blotting is useful when you have a limited amount of sample material, as it requires smaller sample volumes.
- Initial Screening: It is an effective tool for initial screenings to determine the presence or absence of target molecules.
- Versatility: Dot blotting can be adapted for the detection of various biomolecules, including proteins, DNA, RNA, and more.
- Cost-Effective: It is often more cost-effective than quantitative assays, making it suitable for labs with budget constraints.
- Resource-Friendly: Dot blotting requires fewer resources and equipment compared to some other molecular techniques.
Disadvantages of Dot Blotting:
- Lack of Quantification: Dot blotting provides only qualitative data and does not quantify the exact amount of the target molecule.
- Reduced Sensitivity: It may have lower sensitivity compared to more quantitative techniques like ELISA or qPCR.
- Cross-Reactivity: Non-specific binding or cross-reactivity with other molecules can lead to false-positive results.
- Limited Dynamic Range: Dot blotting has a limited dynamic range, making it less suitable for quantifying samples with a wide range of concentrations.
- Limited Specificity: The specificity of dot blotting relies heavily on the quality and specificity of antibodies or probes used.
- Sample Distribution: Achieving uniform dot distribution on the membrane can be challenging, affecting the consistency of results.
- Confirmation Needed: When precise quantification is required, dot blot results should be confirmed with more quantitative methods.
- Background Noise: Non-specific background signals can sometimes interfere with result interpretation.
- Difficulty in Multiplexing: Detecting multiple targets simultaneously (multiplexing) can be challenging in dot blotting.
- Data Variability: The interpretation of dot blot results can be subjective and may vary between individuals.
Limitations of Dot Blotting:
- Qualitative Data: Dot blotting provides only qualitative data, indicating the presence or absence of a target molecule. It does not quantify the exact amount of the target.
- Reduced Sensitivity: It may have lower sensitivity compared to more quantitative techniques like ELISA or quantitative PCR (qPCR), making it less suitable for detecting low-abundance targets.
- Cross-Reactivity: Non-specific binding or cross-reactivity with other molecules can lead to false-positive results, particularly if the specificity of the antibody or probe is not high.
- Limited Dynamic Range: Dot blotting has a limited dynamic range, making it less suitable for quantifying samples with a wide range of concentrations.
- Sample Distribution: Achieving uniform dot distribution on the membrane can be challenging, potentially leading to inconsistency in results.
- Background Noise: Non-specific background signals can sometimes interfere with result interpretation, especially in samples with high background.
- Subjective Interpretation: The interpretation of dot blot results can be subjective and may vary between individuals, leading to potential bias.
- Multiplexing Challenges: Detecting multiple targets simultaneously (multiplexing) can be challenging in dot blotting compared to other techniques like microarrays.
- Limited Quantification: It cannot provide precise quantitative data, which is often required in many research and diagnostic applications.
- Confirmation Needed: When precise quantification is essential, dot blot results should be confirmed with more quantitative methods, which adds an extra step to the analysis.
- Specificity Relies on Probes: The specificity of dot blotting relies heavily on the quality and specificity of the antibodies or probes used. Low-quality probes can result in false results.
- Difficulty in High-Throughput: Dot blotting is not well-suited for high-throughput applications due to the manual nature of dot application and analysis.
Variations and Modern Alternatives:
- Slot Blotting: Slot blotting is similar to dot blotting but uses slots or wells to apply the sample instead of dots. This can provide more consistent sample loading and is often used for quantitative analysis.
- Enzyme-Linked Immunosorbent Assay (ELISA): ELISA is a highly sensitive and quantitative technique that uses antibodies to detect and quantify specific antigens in samples. It is often preferred when precise quantification is required.
- Lateral Flow Assays: Lateral flow assays are rapid diagnostic tests that use capillary action to move a sample along a nitrocellulose membrane where target molecules react with specific antibodies, producing visible test lines. They are commonly used for point-of-care testing.
- Reverse Dot Blot: In reverse dot blotting, known DNA or RNA probes are immobilized on the membrane, and the sample containing complementary sequences is applied. This is used for identifying specific genetic mutations or sequences.
- Microarray Technology: Microarrays consist of thousands of spots, each with a unique probe sequence. They are used for high-throughput analysis of gene expression, genotyping, and other applications. Protein microarrays also allow high-throughput protein profiling.
- Fluorescent Dot Blot: Fluorescent dot blotting uses fluorescently labeled probes and detection reagents to enhance sensitivity and allow for quantitative analysis. It is commonly used for protein detection.
- Bead-Based Assays: Bead-based assays use microspheres or beads coated with specific capture molecules (e.g., antibodies or DNA probes) to detect and quantify multiple targets simultaneously in a liquid-phase assay.
- Real-Time PCR (qPCR): Quantitative PCR (qPCR) is a highly sensitive and quantitative method for amplifying and detecting DNA or RNA sequences. It allows real-time monitoring of the amplification process, enabling accurate quantification.
- Next-Generation Sequencing (NGS): NGS technologies provide comprehensive analysis of DNA or RNA sequences, enabling the detection of multiple targets and providing vast amounts of sequencing data.
- Digital PCR (dPCR): Digital PCR is a highly precise and quantitative method for nucleic acid detection that partitions samples into thousands of individual reactions, allowing absolute quantification of target molecules.
Comparison of Dot Blotting with Modern Techniques:
| Aspect | Dot Blotting | ELISA | qPCR | NGS |
|---|---|---|---|---|
| Type of Technique | Qualitative | Quantitative | Quantitative | Quantitative & Qualitative |
| Target Molecule | Proteins, DNA, RNA | Proteins, DNA, RNA | DNA, RNA | DNA, RNA, Proteins |
| Sensitivity | Low to Moderate | High | High | High |
| Specificity | Medium to High | High | High | High |
| Quantitative | No | Yes | Yes | Yes |
| Multiplexing Capability | Limited | Limited to Moderate | Moderate to High | High |
| Sample Volume Required | Low | Moderate | Low | Variable |
| Speed | Quick | Moderate to Slow | Moderate | Variable (can be slow) |
| Data Output | Qualitative | Quantitative | Quantitative | Quantitative & Qualitative |
| Cost | Low to Moderate | Moderate to High | Moderate to High | High |
| Equipment | Minimal | ELISA reader | qPCR instrument | Specialized sequencing instruments |
| Application Range | Initial Screening | Quantitative Analysis | Quantitative Analysis | Comprehensive Analysis |
| Typical Use Cases | Presence/Absence | Biomarker Detection | Gene Expression, Genotyping | Genomic Sequencing, Transcriptomics, Proteomics |
| Resources Required | Fewer | More | More | More |
FAQs:
1. What is dot blotting used for?
- Dot blotting is used for the qualitative detection of specific molecules (proteins, DNA, RNA, etc.) in a sample. It is often used for initial screenings, assessing the presence or absence of target molecules.
2. How does dot blotting differ from Western blotting?
- Dot blotting is a simplified, qualitative technique that involves applying sample drops directly to a membrane. In contrast, Western blotting is a more complex, quantitative technique that separates proteins by electrophoresis before transferring them to a membrane.
3. What are the limitations of dot blotting?
- Dot blotting provides qualitative, not quantitative, data. It has reduced sensitivity compared to some other methods and can be prone to cross-reactivity or non-specific binding.
4. What are modern alternatives to dot blotting?
- Modern alternatives include ELISA (for quantitative protein detection), qPCR (for nucleic acid quantification), microarrays (for high-throughput gene expression analysis), and NGS (for comprehensive genomic, transcriptomic, and proteomic analysis).
5. When should I choose dot blotting over other techniques?
- Dot blotting is suitable when you have limited sample material, need a quick qualitative assessment, or when resources are limited. It’s a valuable initial screening tool.
6. Can dot blotting be quantitative?
- Dot blotting is primarily qualitative, but it can be semi-quantitative if signal intensity is compared across samples. However, for precise quantification, other methods like ELISA or qPCR are preferred.
7. How can I reduce non-specific binding in dot blotting?
- Proper blocking and choice of blocking agents (e.g., BSA or milk) can reduce non-specific binding. Additionally, optimizing the washing steps can help minimize background noise.
8. What are the advantages of ELISA over dot blotting for protein detection?
- ELISA offers quantitative results, higher sensitivity, and greater specificity due to the use of paired antibodies. It is preferred when precise quantification and specificity are required.
9. Is dot blotting suitable for detecting genetic mutations?
- Dot blotting can be adapted for detecting genetic mutations using reverse dot blot techniques, where known probes are immobilized on the membrane, and sample DNA is applied to identify specific mutations.
10. Can dot blotting be used for gene expression analysis?
- While dot blotting is not commonly used for gene expression analysis, it can be adapted for qualitative assessment of specific RNA transcripts. However, qPCR and microarray technologies are more commonly used for this purpose.
Conclusion:
In conclusion, dot blotting is a versatile and valuable technique in molecular biology and diagnostics. It allows for the qualitative detection of specific molecules, including proteins, DNA, and RNA, making it useful for initial screenings, assessments of target molecule presence, and various research applications. However, dot blotting has limitations, such as its qualitative nature and potential for non-specific binding, which may require confirmation with more quantitative methods when precise quantification is essential.
In the ever-evolving field of molecular biology, researchers have access to a wide range of modern alternatives, including ELISA, qPCR, microarrays, and next-generation sequencing (NGS), which offer greater sensitivity, quantification capabilities, and multiplexing options. The choice between dot blotting and these modern techniques depends on the specific research goals, available resources, and the need for qualitative or quantitative data.