Droplet Digital PCR (ddPCR) is a highly precise and sensitive method for absolute quantification of nucleic acids. It partitions a sample into thousands of droplets, with PCR amplification occurring in each droplet. This technique is used for detecting rare mutations, measuring gene expression, and quantifying viral load with high accuracy.
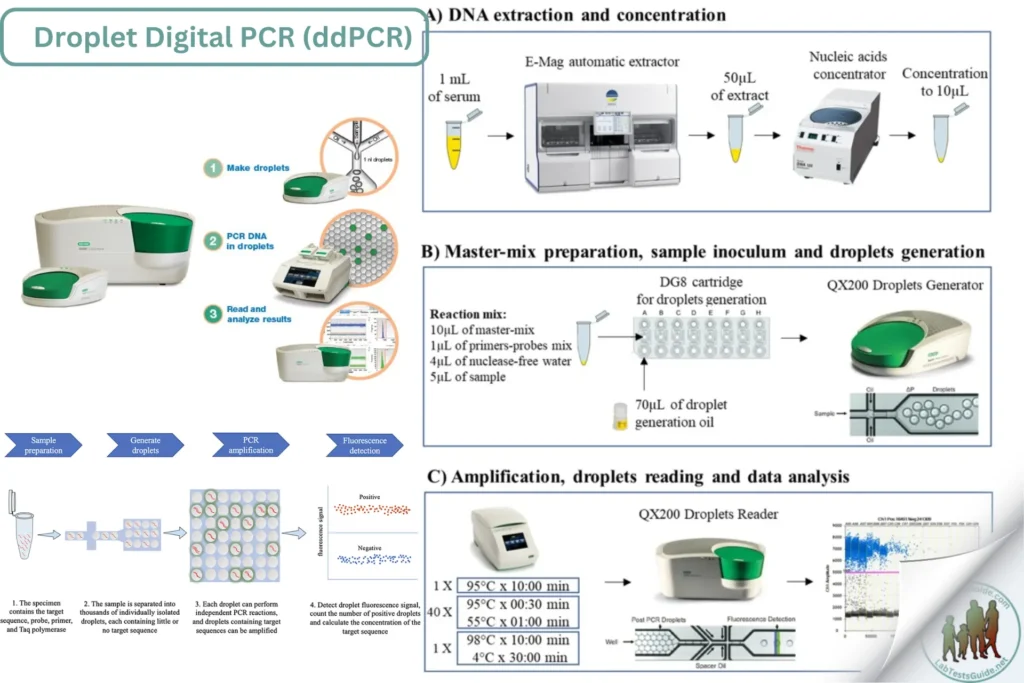
Methodology for Droplet Digital PCR (ddPCR) in Clinical Laboratory
Introduction
Droplet Digital PCR (ddPCR) is a highly precise and sensitive method for absolute quantification of nucleic acids. It partitions a sample into thousands of droplets, with PCR amplification occurring in each droplet. This technique is used for detecting rare mutations, measuring gene expression, and quantifying viral load with high accuracy.
Principle
ddPCR involves partitioning the sample into thousands of nanoliter-sized droplets. Each droplet acts as an individual PCR reaction. After PCR amplification, the droplets are analyzed for fluorescence to determine the presence of the target DNA, allowing for absolute quantification based on the number of positive droplets.
Specimen Requirements
- Type of specimen: Blood, plasma, serum, tissue samples, swabs, or other biological fluids
- Volume of specimen required: Typically 10-50 µL
- Collection method: Standard clinical procedures for sample collection (e.g., venipuncture, swabs)
- Handling and storage requirements: Store samples at -80°C for long-term storage; avoid repeated freeze-thaw cycles
- Stability of the specimen: DNA is generally stable, but follow specific guidelines for sample type
Reagents and Materials
- DNA extraction kit
- ddPCR master mix
- Specific primers and probes for the target DNA
- Droplet generation oil
- Nuclease-free water
- Positive and negative control DNA
Equipment
- Droplet generator (e.g., Bio-Rad QX200)
- ddPCR thermal cycler
- Droplet reader
- Microcentrifuge
- Vortex mixer
- Micropipettes and RNase/DNase-free tips
- ddPCR consumables (cartridges, gaskets, and plates)
- Biosafety cabinet (for handling DNA)
Procedure
DNA Extraction:
- Extract DNA from the sample using a DNA extraction kit following the manufacturer’s instructions.
- Quantify the extracted DNA using a spectrophotometer or fluorometer.
- Assess DNA quality and integrity using gel electrophoresis or a bioanalyzer (optional).
Droplet Generation:
- Prepare the ddPCR reaction mix:
- DNA template: 1-10 ng
- ddPCR master mix: 1X
- Forward and reverse primers: 0.2-0.5 µM each
- Probe: 0.1-0.2 µM
- Nuclease-free water to the final volume (usually 20-25 µL)
- Load the reaction mix and droplet generation oil into the droplet generator cartridges.
- Use the droplet generator to partition the sample into droplets.
PCR Amplification:
- Transfer the droplets to a PCR plate.
- Set up the PCR cycling conditions:
- Initial denaturation: 95°C for 10 minutes
- Amplification (40-45 cycles):
- Denaturation: 95°C for 30 seconds
- Annealing/Extension: 55-60°C for 1-2 minutes
- Final extension: 72°C for 5 minutes (if required)
- Perform PCR amplification in the ddPCR thermal cycler.
Droplet Reading:
- After PCR amplification, transfer the droplets to the droplet reader.
- Analyze the droplets for fluorescence to determine the number of positive and negative droplets.
- Use the droplet reader software to count the number of positive droplets and calculate the absolute quantity of target DNA.
Quality Control
- No-Template Controls (NTCs): Include NTCs to check for contamination.
- Positive and Negative Controls: Validate the efficiency and specificity of the assay.
- Droplet Generation Efficiency: Ensure proper droplet generation to avoid false negatives or positives.
- Instrument Calibration: Regularly calibrate the ddPCR instruments according to the manufacturer’s guidelines.
Calculation and Interpretation
- Absolute Quantification: Calculate the absolute number of target DNA molecules in the sample based on the proportion of positive droplets using Poisson statistics.
- Result Interpretation: Compare results with established reference ranges or clinical guidelines for accurate interpretation.
- Data Analysis Software: Use the manufacturer’s software for data analysis and visualization.
Limitations
- Sample Quality: Poor-quality DNA can affect droplet generation and amplification efficiency.
- Complexity: ddPCR is more complex and time-consuming compared to traditional PCR.
- Cost: ddPCR requires specialized equipment and reagents, making it more expensive.
Safety Precautions
- PPE: Use appropriate PPE (gloves, lab coat, safety goggles) when handling samples and reagents.
- Clean Environment: Work in a DNase/RNase-free environment to prevent contamination.
- Waste Disposal: Dispose of waste according to local regulations and institutional guidelines.
References
- Huggett, J. F., Foy, C. A., Benes, V., Emslie, K., Garson, J. A., Haynes, R., … & Saunders, N. A. (2013). The digital MIQE guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments. Clinical Chemistry, 59(6), 892-902.
- ddPCR instrument manufacturer’s guidelines and protocols.
- Molecular Cloning: A Laboratory Manual (Fourth Edition) by Michael R. Green and Joseph Sambrook.
- Clinical guidelines and standards from institutions such as the American Society for Microbiology (ASM) and the Clinical and Laboratory Standards Institute (CLSI).
This format provides a structured approach to conducting ddPCR in clinical laboratories, focusing on accurate absolute quantification of nucleic acids, quality control, and safety precautions for reliable results.