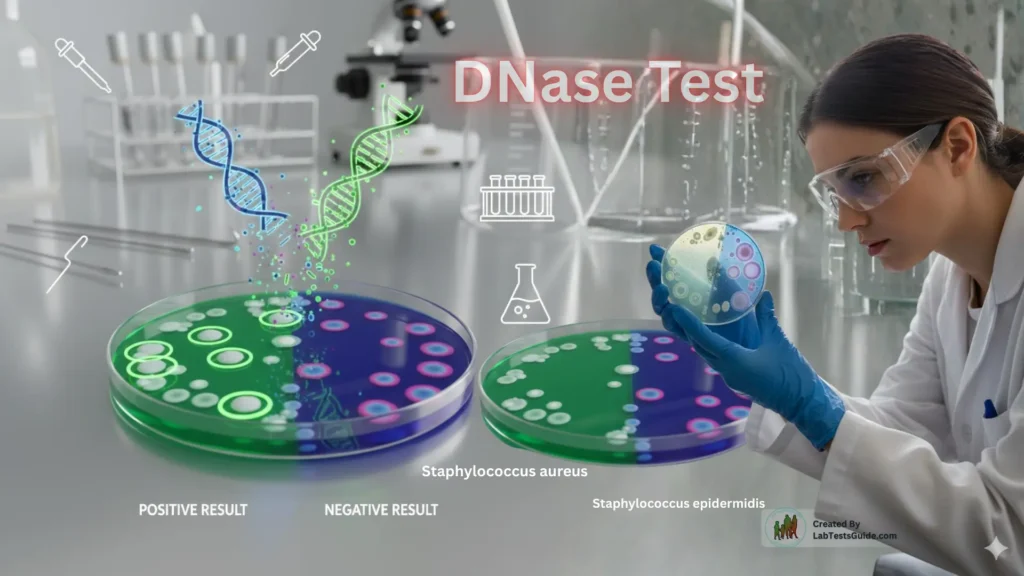
The DNase Test is a widely used biochemical test in microbiology to detect the ability of bacteria to produce deoxyribonuclease (DNase) — an enzyme that breaks down DNA. It is especially valuable for differentiating Staphylococcus aureus from other staphylococci, identifying Gram-negative pathogens, and supporting clinical diagnosis.
This complete guide covers everything you need: principle, agar composition, procedure, result interpretation, quality control, uses, limitations, and FAQs.
What is the DNase Test?
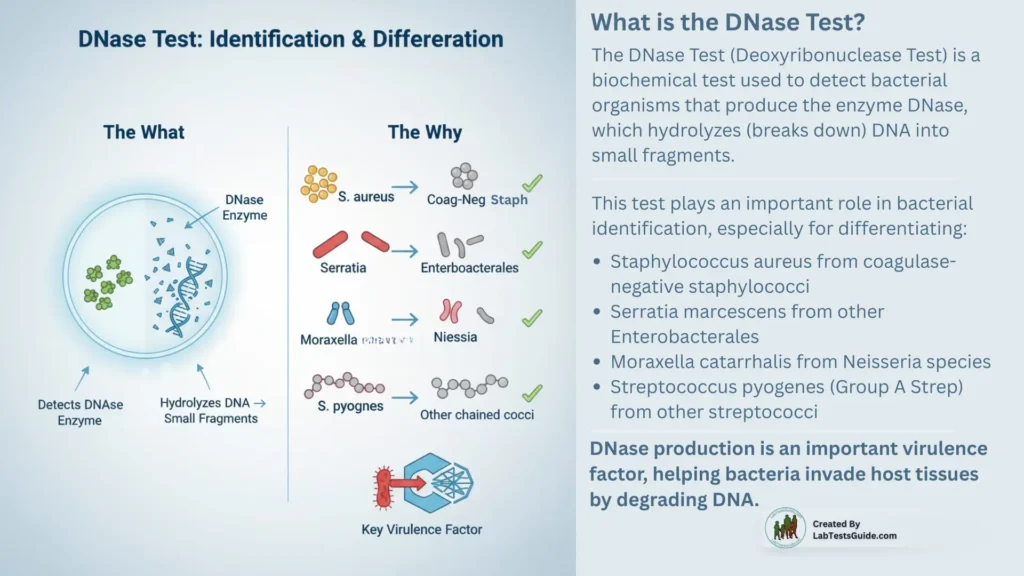
The DNase Test (Deoxyribonuclease Test) is a biochemical test used to detect bacterial organisms that produce the enzyme DNase, which hydrolyzes (breaks down) DNA into small fragments. This test plays an important role in bacterial identification, especially for differentiating:
- Staphylococcus aureus from coagulase-negative staphylococci
- Serratia marcescens from other Enterobacterales
- Moraxella catarrhalis from Neisseria species
- Streptococcus pyogenes (Group A Strep) from other streptococci
DNase production is an important virulence factor, helping bacteria invade host tissues by degrading DNA.
Why the DNase Test Matters
- Helps confirm suspected S. aureus when coagulase results are unclear
- Provides rapid differentiation in routine bacteriology
- Useful in teaching labs and clinical microbiology workflows
- Works with both dye-based DNase agar plates and the classical HCl flooding method
Historical Background
The DNase test was first developed in 1957 by Jeffries et al., who used an acid-precipitation method to detect DNA hydrolysis. Their method relied on flooding DNase agar with 1N HCl, which precipitates intact DNA, producing an opaque background.
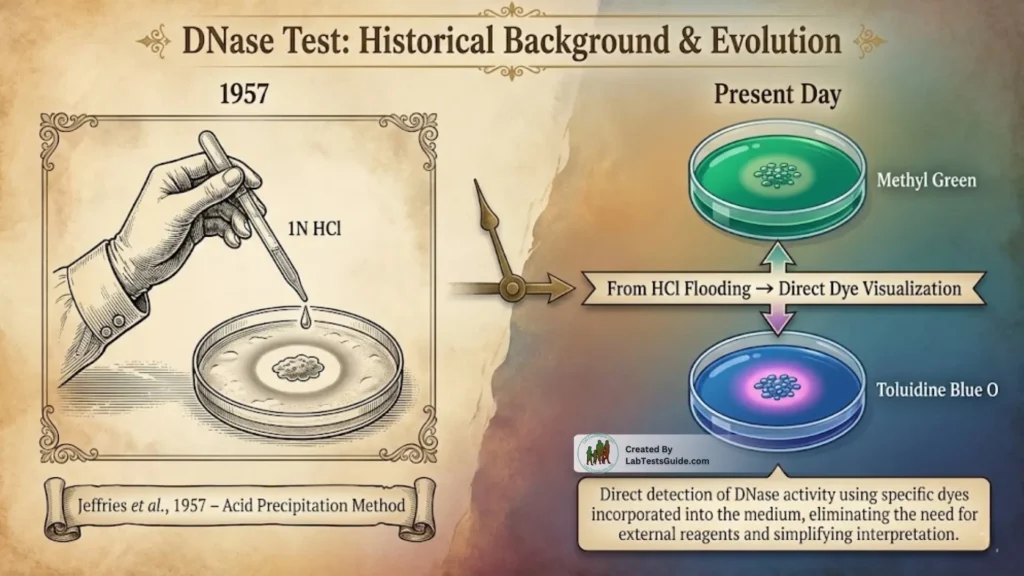
Areas where DNA was hydrolyzed remained clear, forming a visible halo.
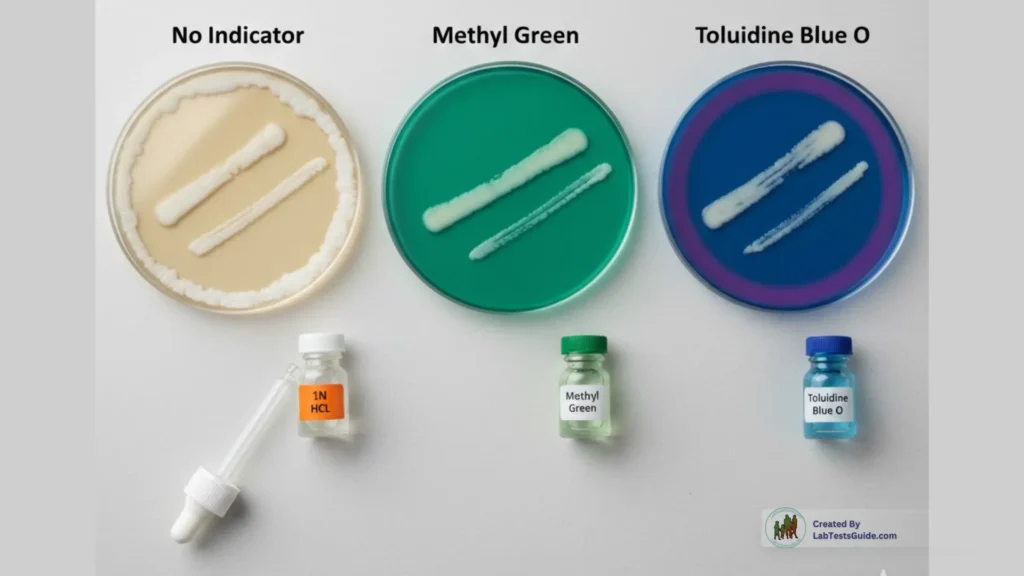
Later advancements introduced indicator dyes such as:
- Methyl Green (forms a DNA–dye complex; turns colorless when DNA is degraded)
- Toluidine Blue O (produces a pink–purplish halo around DNase-positive colonies)
These dye-based media eliminated the need for HCl and allowed direct, non-destructive visualization of test results.
Principle of the DNase Test
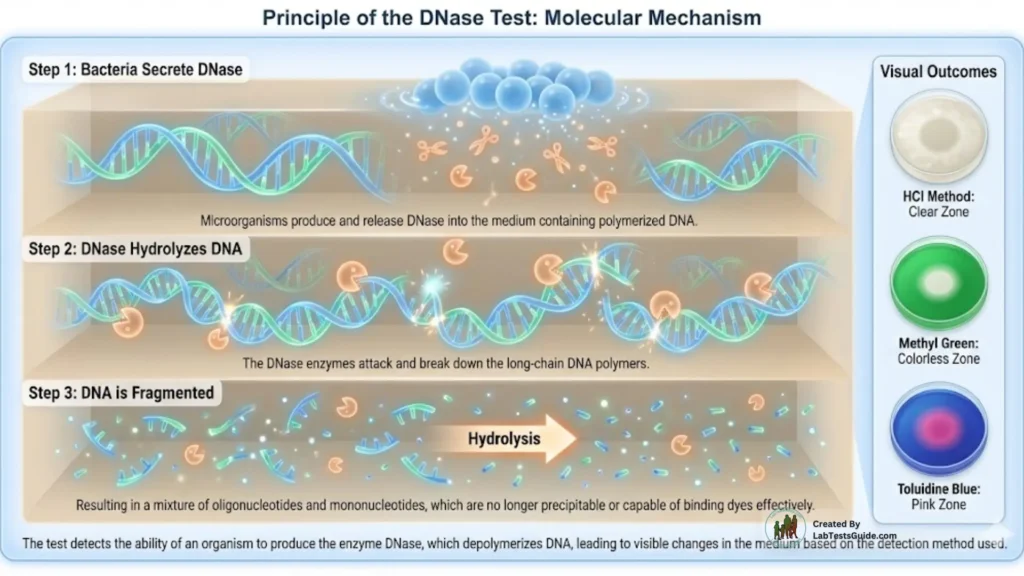
DNase-positive organisms hydrolyze DNA in the medium.
When HCl is added, it precipitates unhydrolyzed DNA, creating a cloudy, opaque background.
Clear zones around the colonies indicate DNA degradation, meaning the organism produces DNase.
Some formulations include methyl green, which forms a colored complex with DNA and becomes clear when DNA is hydrolyzed.
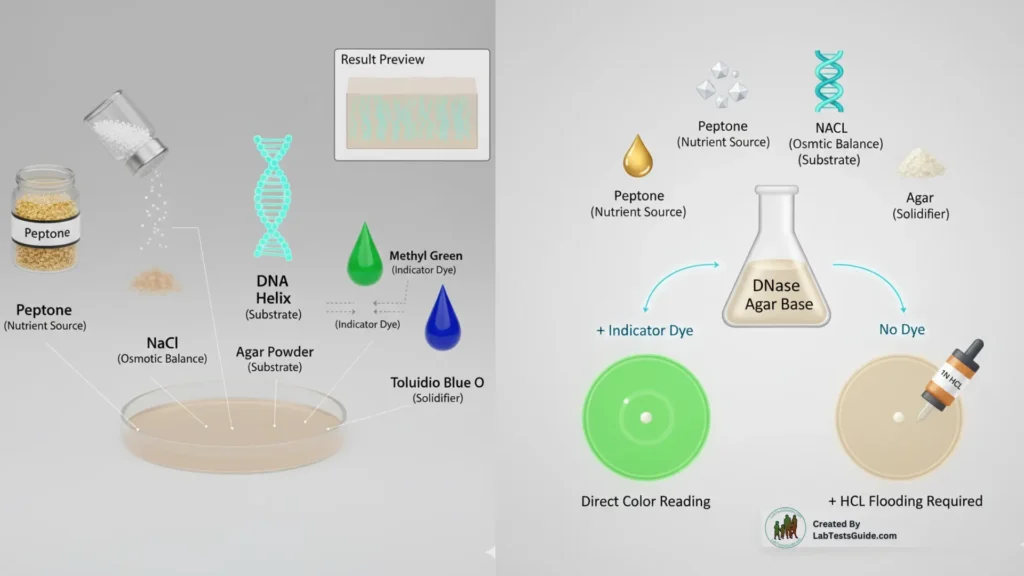
Composition of DNase Agar
Typical DNase agar contains:
- Peptone – Nutrient source
- Sodium Chloride – Maintains osmotic balance
- DNA (substrate) – Detects DNase activity
- Agar – Solidifies the medium
- Indicator (optional) – Methyl green or toluidine blue
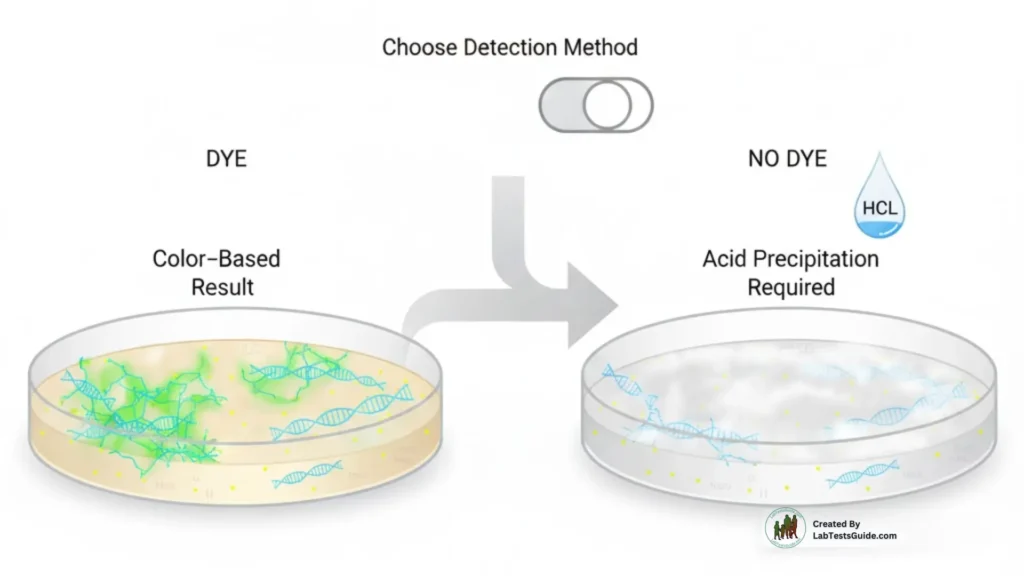
With indicators → color-based reading
Without indicators → requires HCl flooding post-incubation
Types of DNase Test Methods:

1. DNase Agar Plate Method (Without Indicator)
- After incubation, 1N HCl is added.
- HCl precipitates intact DNA → Cloudy background.
- DNase-positive organisms show a clear halo.
2. DNase Agar with Methyl Green
- No need for HCl.
- Intact DNA binds methyl green (green color).
- Hydrolysis → colorless or pale zone.
3. DNase Agar with Toluidine Blue O
- DNA hydrolysis changes dye color from blue to pink.

Materials Required:
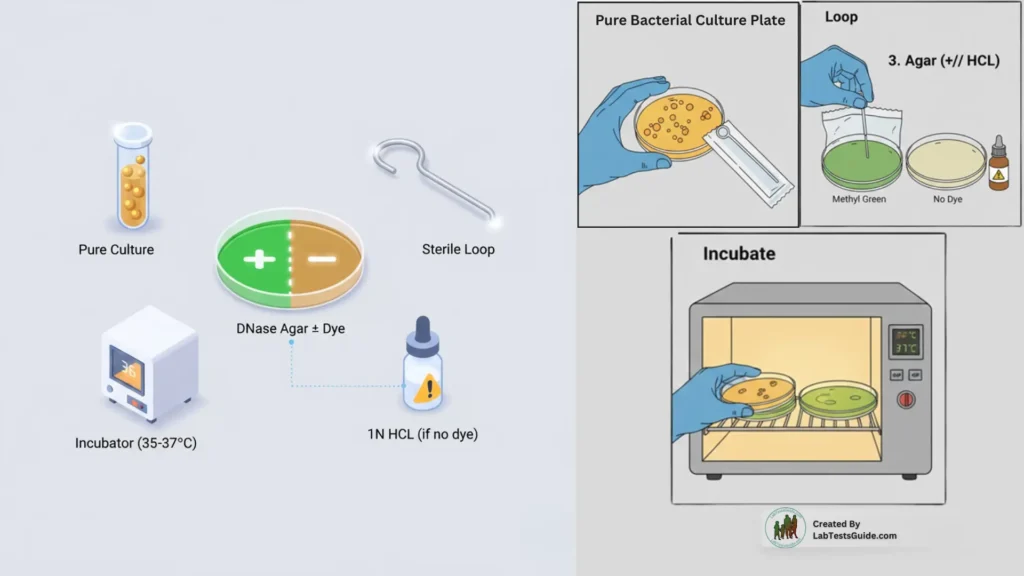
- DNase agar (with or without indicator)
- Sterile loop
- Incubator (35–37°C)
- HCl (if using indicator-free agar)
- Pure bacterial culture
Preparation of DNase Agar Media

- Weighing and Mixing
- Measure 42 grams of dehydrated DNase agar powder (or use lab-prepared media).
- Add it to 1000 mL of pure distilled or deionized water in a clean beaker.
- Dissolving the Medium
- Heat the suspension while stirring or agitating until it comes to a boil and the medium is completely dissolved.
- Sterilization
- Autoclave the dissolved medium at 121°C (15 lbs pressure) for 15 minutes.
- Cooling
- After autoclaving, allow the medium to cool to approximately 40–45°C.
- Adding Indicators (if required)
- If using methyl green or toluidine blue/O, add 0.1 gram to the cooled medium before sterilization for proper DNA detection.
- Pouring Plates
- Pour the sterile medium into sterile Petri plates under aseptic conditions.
- Allow the plates to solidify before use.
Procedure of DNase Test

- Preparation of DNase Agar Plate
- Use DNase agar medium (nutrient agar with DNA and methyl green or toluidine blue).
- The indicator binds to DNA, producing a colored complex. DNase activity will degrade DNA and change the color around bacterial growth.
- Inoculation
- Streak the test organism in a straight line or spot-inoculate onto the DNase agar plate.
- Include a positive control (known DNase-producing bacterium, e.g., Staphylococcus aureus) and a negative control (DNase-negative bacterium, e.g., Staphylococcus epidermidis).
- Incubation
- Incubate the plate at 35–37°C for 24–48 hours.
- Conditions should be optimal for bacterial growth.
- Detection of DNase Activity
- Direct observation method (with methyl green):
- A clear zone around the bacterial growth indicates DNA degradation (positive DNase activity).
- No change in color indicates a negative result.
- Flooding method (optional):
- Flood the plate with 1N HCl after incubation.
- HCl precipitates intact DNA, turning the agar opaque.
- Clear zones around colonies indicate DNase-positive organisms.
- Direct observation method (with methyl green):
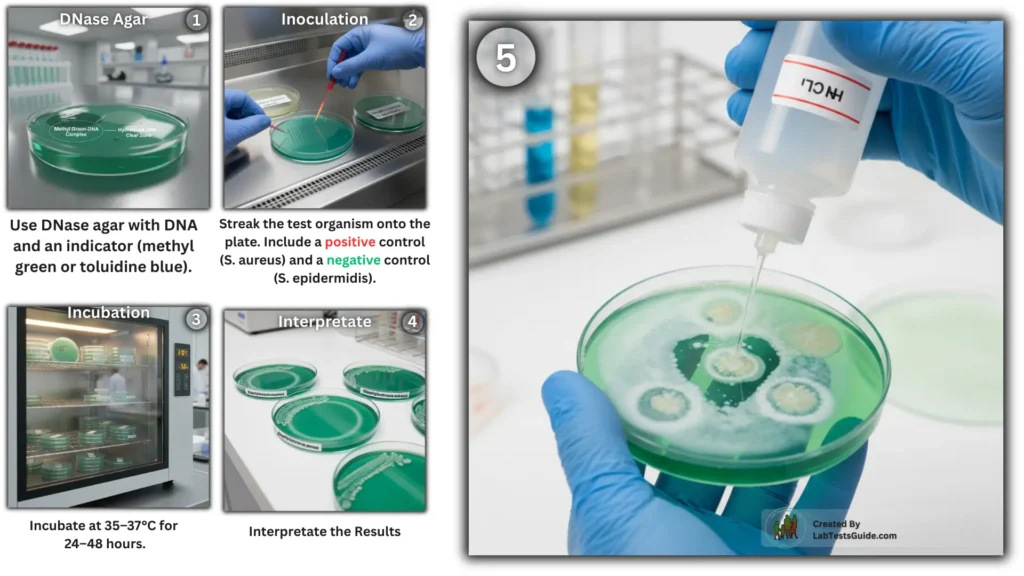
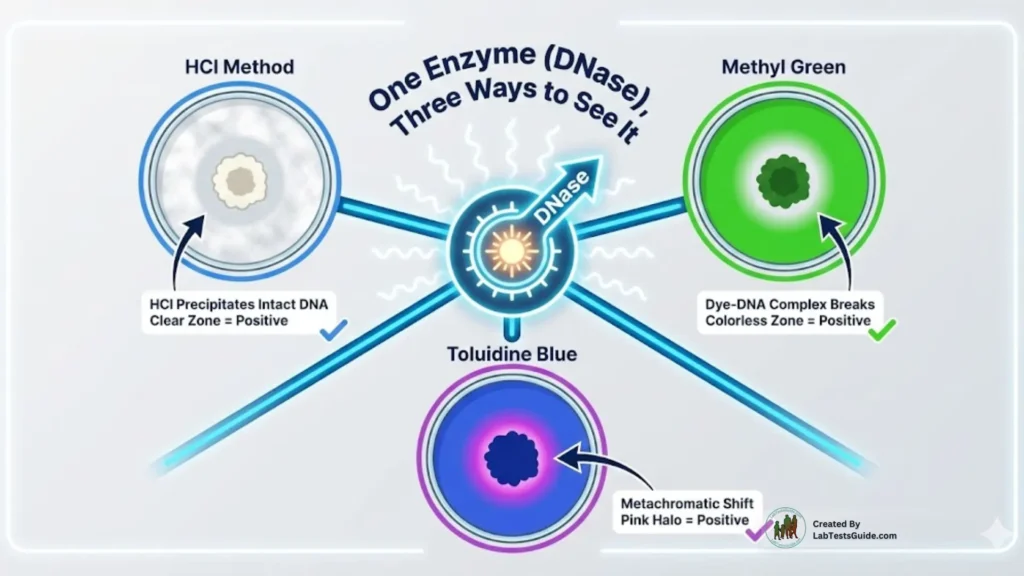
Result Interpretation
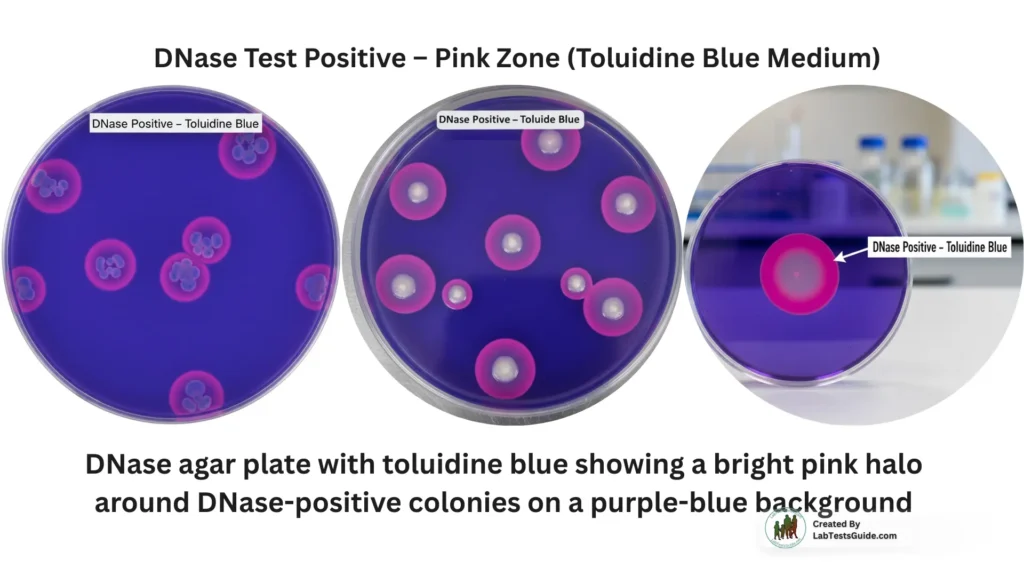
DNase Test Positive
- Clear zone around growth (without indicator)
- Colorless zone (methyl green)
- Pink zone (toluidine blue)
Examples of DNase-positive organisms:
- Staphylococcus aureus
- Serratia marcescens
- Moraxella catarrhalis
- Campylobacter jejuni
DNase Test Negative
- No clear or colorless zone
- Medium remains uniform around colonies
Examples:
- Staphylococcus epidermidis
- Many Streptococci
- Most Enterobacterales except Serratia





Clinical Importance
The DNase test is essential because DNase production is often associated with virulence, helping differentiate clinically significant pathogens. It supports identification in:
- Skin and soft tissue infections
- Respiratory pathogens
- Hospital-acquired infections
Quality Control
Use standard ATCC strains:
| Organism | Expected Result |
|---|---|
| Staphylococcus aureus ATCC 25923 | Positive |
| Serratia marcescens ATCC 8100 | Positive |
| Staphylococcus epidermidis ATCC 12228 | Negative |
QC should be done whenever:
- A new batch of media is prepared
- A new shipment is received
- Results appear questionable
Common Errors & How to Avoid Them
- Over-incubation → false positives
- Too heavy inoculum → masking zones
- Dry plates → inaccurate results
- Using expired HCl → poor DNA precipitation
Applications of the DNase Test
- Differentiation of staphylococci
- Identification of Serratia species
- Characterization of Gram-negative rods
- Virulence and pathogenicity assessment
- Supportive tool in clinical microbiology workflows
Limitations
- Some organisms produce weak DNase → indistinct results
- Requires fresh media for accurate visibility
- HCl flooding sometimes diffuses colonies
- Should be used along with other biochemical tests for final identification
DNase positive bacteria
DNase negative bacteria
DNase test results
DNase agar with methyl green
DNase agar with toluidine blue
HCl method DNase test
DNase quality control strains
Microbiology biochemical tests
DNase positive bacteria
DNase negative bacteria
DNase test results
DNase agar with methyl green
DNase agar with toluidine blue
HCl method DNase test
DNase quality control strains
Microbiology biochemical tests
⚠️ Disclaimer:
The content on LabTestsGuide.com is for informational and educational purposes only. We do not guarantee the accuracy, completeness, or timeliness of the information provided. Always consult qualified healthcare professionals for medical advice, diagnosis, or treatment. LabTestsGuide.com is not liable for any decisions made based on the information on this site.